Welcome to ALGAEpath 2.0
A free Chlamydomonas reinhardtii gene expression database
Alright let's go
AlgaePath 2.0 is a web-based database integrating gene information, biological pathways, and next-generation sequencing (NGS) datasets derived from Chlamydomonas reinhardtii and Neodesmus sp. UTEX 2219-4. By using Gene Search, Pathway Search, DEGs Search,
Enrichment Analysis, and Co-expression Analysis in AlgaePath, users can effectively identify transcript abundance profiles and compare variations among different conditions in a pathway map.

Provides basic gene information as well as the expression level data under different conditions.
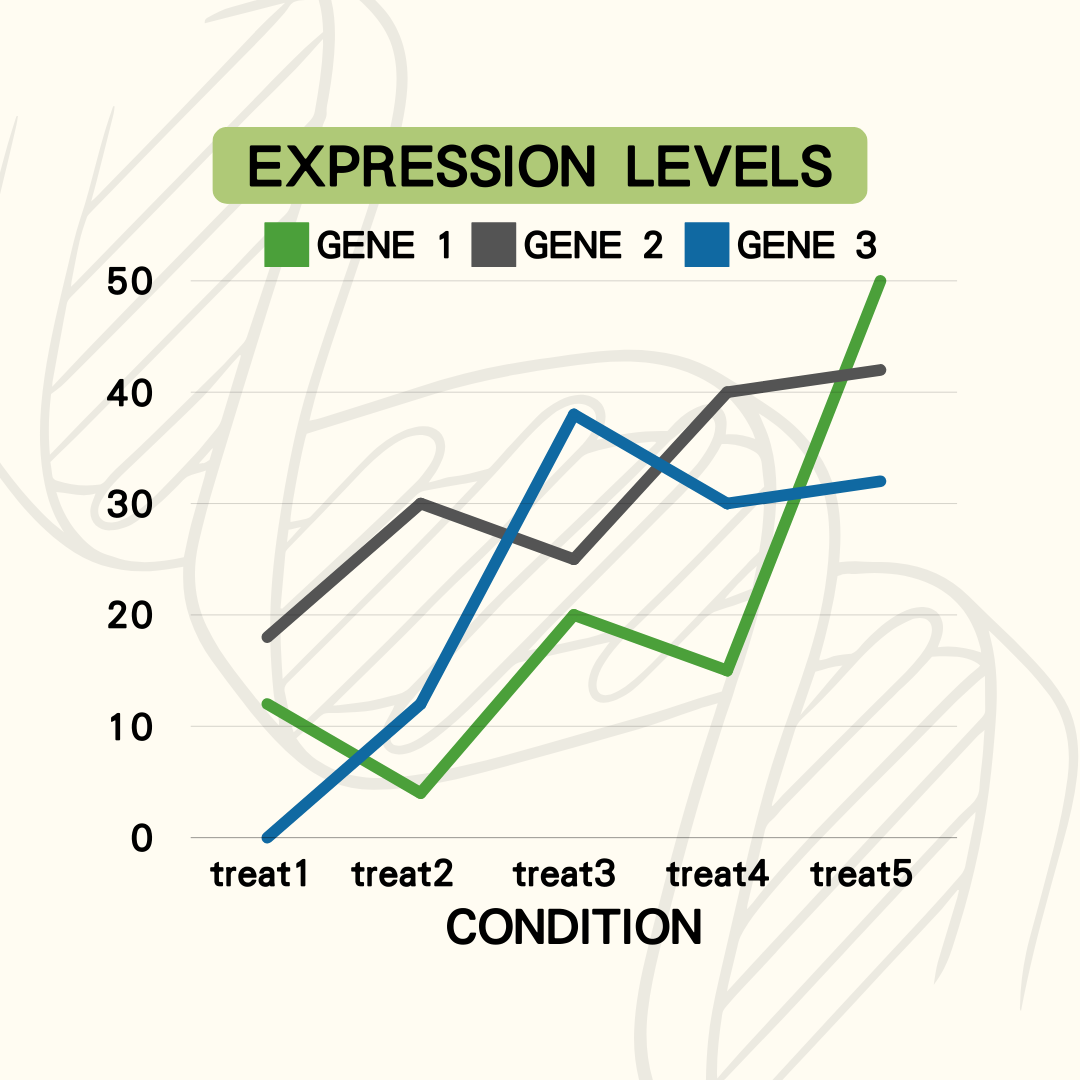
Displays gene expression levels across selected conditions using line charts and heatmaps, based on a gene set.
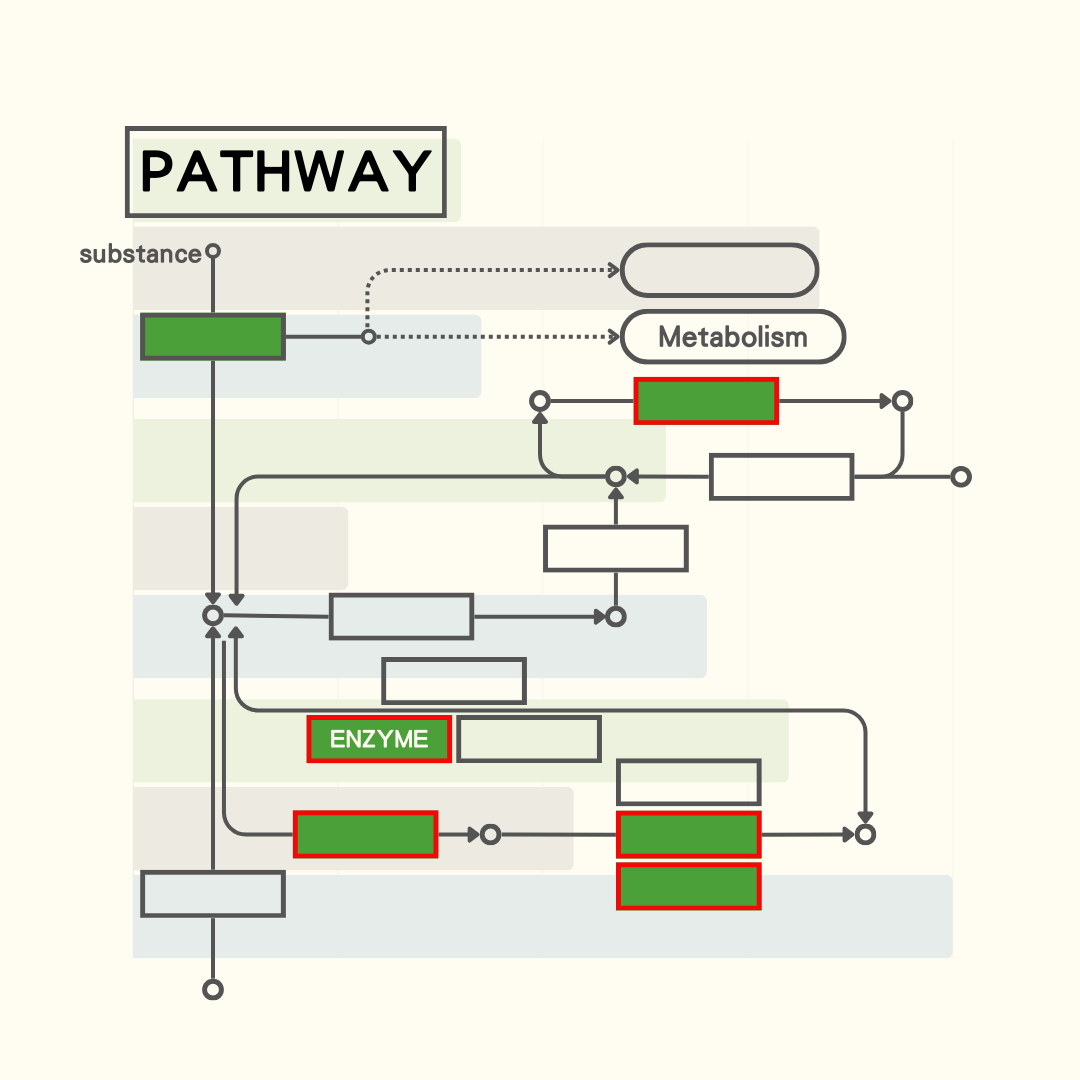
Provides basic information on various KEGG pathways and gene expression data.
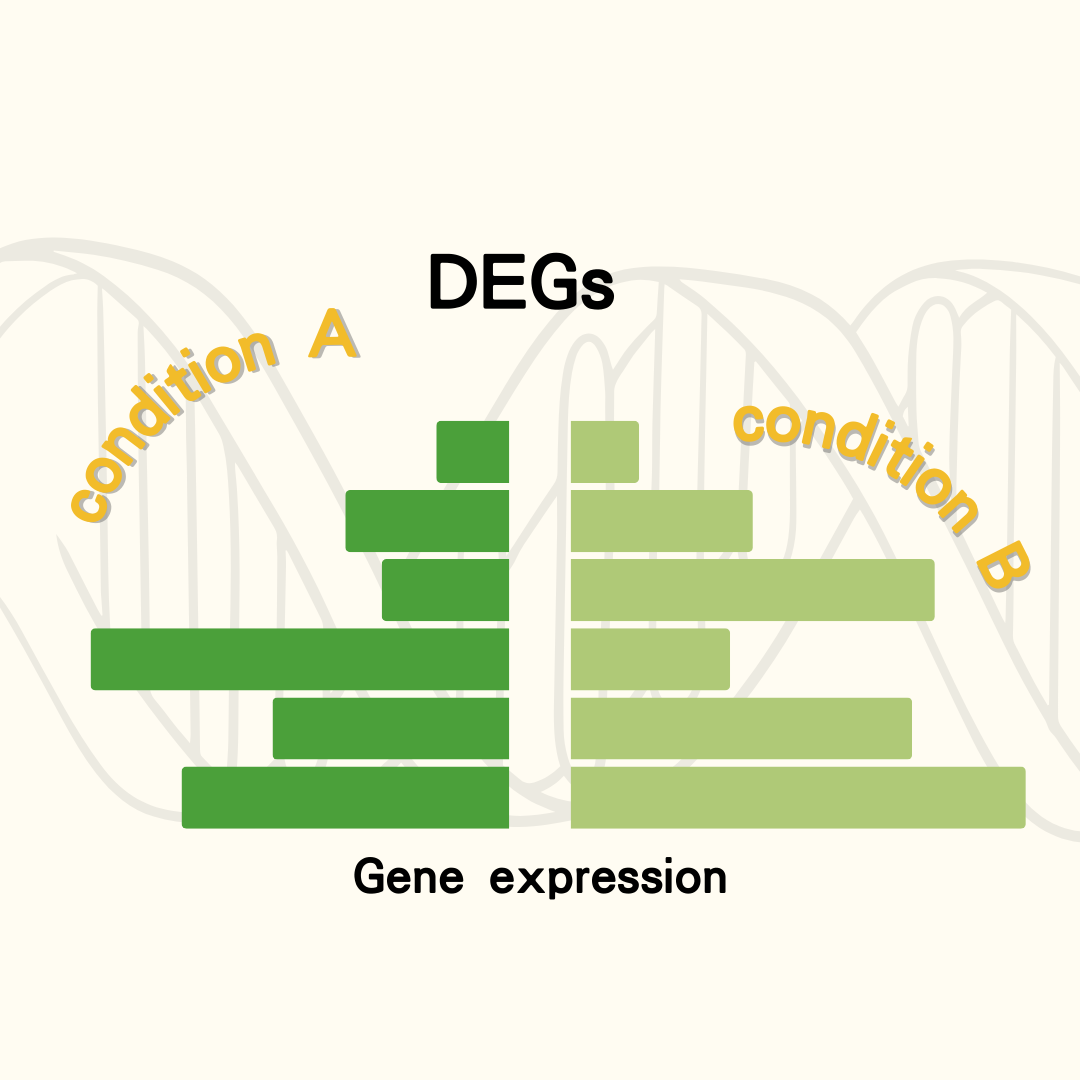
Provides differentially expressed genes between two customized conditions.

Provides information on pathways and Gene Ontology (GO), along with transcript abundance levels of genes under different conditions.
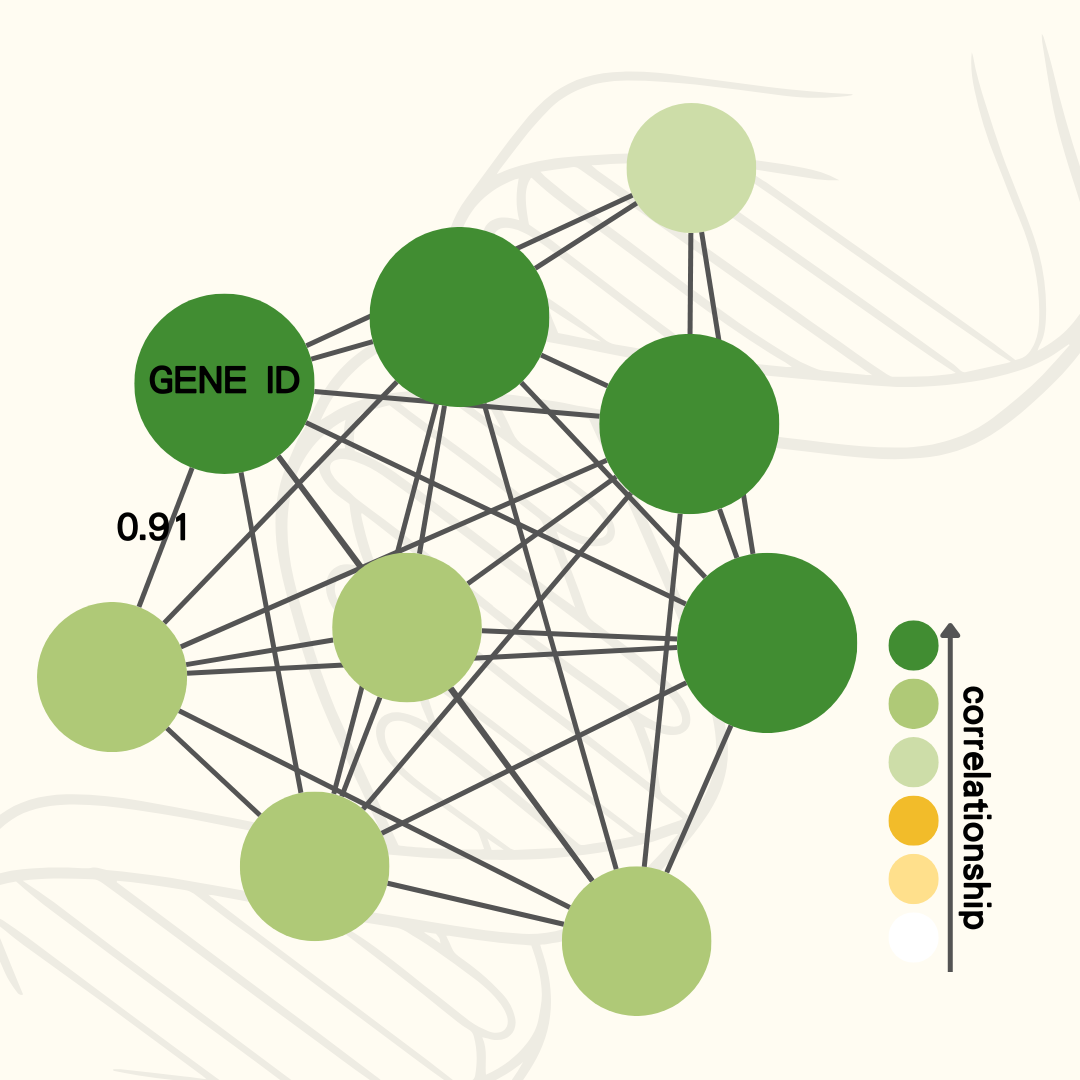
Co-expression genes are provided based on Pearson's correlation coefficient (PCC) or Spearman's rank correlation coefficient.