
 |
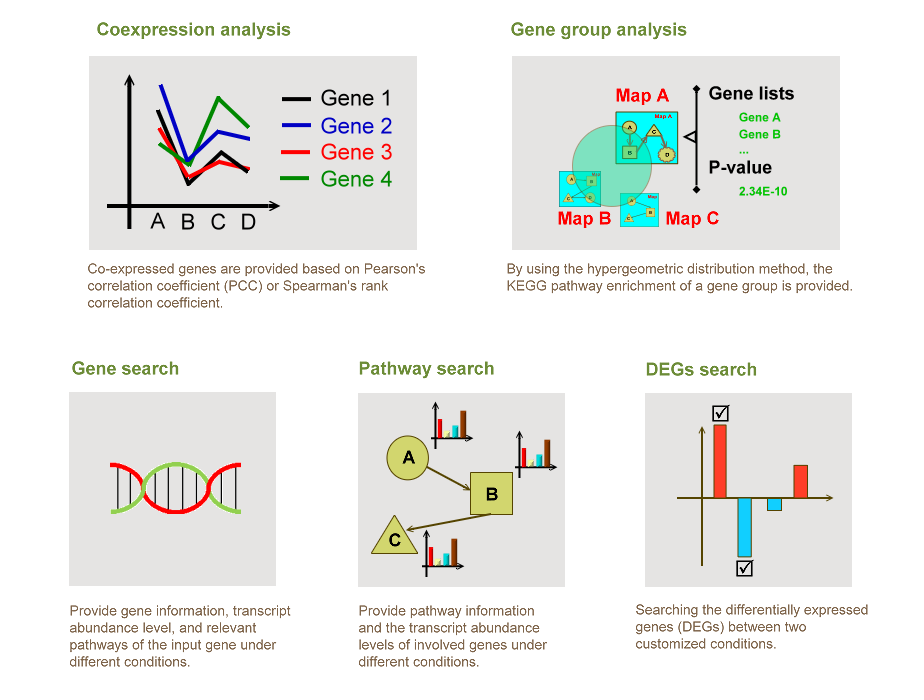
AlgaePath is a web-based database integrating gene information, biological pathways, and next-generation sequencing (NGS) datasets derived from Chlamydomonas reinhardtii and Neodesmus sp. UTEX 2219-4. By using Gene Search, Pathway Search, DEGs Search, Gene Group Analysis, and Co-expression Analysis in AlgaePath, users can effectively identify transcript abundance profiles and compare variations among different conditions in a pathway map.
|
|||
 |
|||